Clinvar citation information
Home » Trending » Clinvar citation informationYour Clinvar citation images are ready in this website. Clinvar citation are a topic that is being searched for and liked by netizens now. You can Get the Clinvar citation files here. Find and Download all royalty-free photos.
If you’re looking for clinvar citation images information connected with to the clinvar citation interest, you have pay a visit to the right blog. Our website frequently gives you suggestions for refferencing the maximum quality video and image content, please kindly surf and find more informative video content and graphics that fit your interests.
Clinvar Citation. Check through this pubmed listing to locate citations/papers on individual ncbi services of your interest. For the blast® services, use these blast references. •consider a “literature only” submission. Clinvar data dictionary, august 4, 2017 page 5 variationrelease xml:
 NCBI Insights ClinVar Unveils New, More Intuitive From ncbiinsights.ncbi.nlm.nih.gov
NCBI Insights ClinVar Unveils New, More Intuitive From ncbiinsights.ncbi.nlm.nih.gov
Clinvar is a public database for clinical laboratories, researchers, expert panels, and others to share their interpretations of variants with their evidence. Aggregating data from many groups in a single database allows comparison of interpretations, providing transparency into the concordance or discordance of interpretations. If you know of citations for this variation, please consider submitting that information to clinvar. Use the citation link on the right side of the pmc view of this article to obtain the citation in the desired format. Gdv showing ‘clinvar variants with precise endpoints’ track next to ncbi human gene annotation. The reason for the remaining 25% was undetermined because no citation in clinvar or the variant was not found in the citation (column n in the supplementary table s1).
Clinvar has a broad scope and includes interpretations of variants in any region of the human genome, including mitochondria.
By the time of submission (clinvar february 2019 release) simple clinvar contains 493,240 genetic. If you know of citations for this variation, please consider submitting that information to clinvar. Check through this pubmed listing to locate citations/papers on individual ncbi services of your interest. Aggregating data from many groups in a single database allows comparison of interpretations, providing transparency into the concordance or discordance of interpretations. The database holds 600,000 submitted records from 1,000 submitters, representing 430,000 unique variants. Clinvar has a broad scope and includes interpretations of variants in any region of the human genome, including mitochondria.

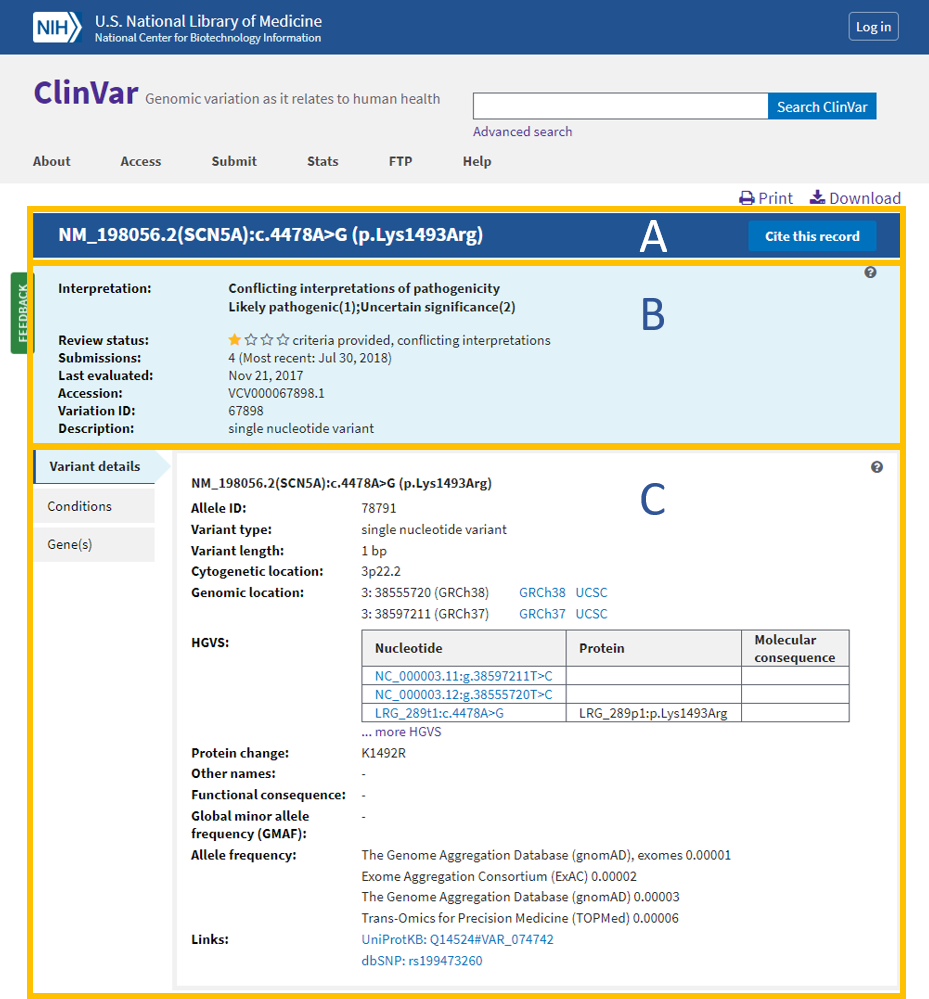
The bottom of the new variation page displays a table of all citations reported to clinvar for the variant with links to pubmed, except for citations provided as assertion criteria and citations that are submitted specifically for the disease (not the variant). Use the citation link on the right side of the pmc view of this article to obtain the citation in the desired format. Want to learn more about who submits to clinvar? The subject of this data analysis experiment is a mashup of two datasets, clinvar and the genetic testing registry.please see the linked blog posts for a detailed introduction to these datasets, as well as their technical details and links to formal documentation. This should be used only when there is no database id for the publication and no url.
 Source: researchgate.net
Source: researchgate.net
Clingen is organizing variant curation expert panels (vceps) to develop specifications to the acmg/amp guidelines for genes or diseases of interest, interpret variants according to these guidelines, and publish the expert interpretations through the publicly available clinvar database. Check through this pubmed listing to locate citations/papers on individual ncbi services of your interest. Citing literature number of times cited according to crossref: • consider batching several citations in one message. •if not, email us at clinvar@ncbi.nlm.nih.gov to ask us to add the citation to the variant.
 Source: ncbi.nlm.nih.gov
Source: ncbi.nlm.nih.gov
Clinvar encourages submissions of variants reviewed by expert panels, as expert consensus confers a high standard. Clinvar data dictionary, august 4, 2017 page 5 variationrelease xml: Citing literature number of times cited according to crossref: Citing the ncbi internet site and individual web pages and records: Public archive of relationships among sequence variation and human phenotype
 Source: researchgate.net
Source: researchgate.net
If you know of citations for this variation, please consider submitting that information to clinvar. Submissions may come from clinical providers providing their own interpretation of the variant (�provider interpretation�) or from groups such as patient registries that primarily provide. Triggered by submitting without a query or with the keyword “clinvar”, it will yield summary statistics of the entire clinvar database. The subject of this data analysis experiment is a mashup of two datasets, clinvar and the genetic testing registry.please see the linked blog posts for a detailed introduction to these datasets, as well as their technical details and links to formal documentation. Want to learn more about who submits to clinvar?
 Source: researchgate.net
Source: researchgate.net
Aggregating data from many groups in a single database allows comparison of interpretations, providing transparency into the concordance or discordance of interpretations. •consider a “literature only” submission. This should be used only when there is no database id for the publication and no url. • consider batching several citations in one message. Clinvar encourages submissions of variants reviewed by expert panels, as expert consensus confers a high standard.
 Source: ncbi.nlm.nih.gov
Source: ncbi.nlm.nih.gov
You have a few options to add the pmid to clinvar •if your vcep will review and submit this variant, include the pmid in your submitted record. Use the citation link on the right side of the pmc view of this article to obtain the citation in the desired format. Variants in clinvar may be of any length or type, ranging from single nucleotide substitutions and small insertions/deletions to copy number changes and cytogenetic rearrangements. Gdv showing ‘clinvar variants with precise endpoints’ track next to ncbi human gene annotation. Triggered by submitting without a query or with the keyword “clinvar”, it will yield summary statistics of the entire clinvar database.
 Source: ncbiinsights.ncbi.nlm.nih.gov
Source: ncbiinsights.ncbi.nlm.nih.gov
Tracks are color coded for quick and easy interpretation. Clinvar is a public database for clinical laboratories, researchers, expert panels, and others to share their interpretations of variants with their evidence. There are no citations in clinvar for this variation. Public archive of relationships among sequence variation and human phenotype Variants in clinvar may be of any length or type, ranging from single nucleotide substitutions and small insertions/deletions to copy number changes and cytogenetic rearrangements.
 Source: ncbiinsights.ncbi.nlm.nih.gov
Source: ncbiinsights.ncbi.nlm.nih.gov
•consider a “literature only” submission. Clinvar is a public database for clinical laboratories, researchers, expert panels, and others to share their interpretations of variants with their evidence. Users can leverage this information in their search of the literature for a variant. Clinvar recently started accepting submissions that are focused primarily on providing phenotypic information for individuals who have had genetic testing. The database holds 600,000 submitted records from 1,000 submitters, representing 430,000 unique variants.
 Source: researchgate.net
Source: researchgate.net
• consider batching several citations in one message. Aggregating data from many groups in a single database allows comparison of interpretations, providing transparency into the concordance or discordance of interpretations. From the front page of simple clinvar the user can submit three types of queries: Want to learn more about who submits to clinvar? Public archive of relationships among sequence variation and human phenotype
 Source: researchgate.net
Source: researchgate.net
In its first five years, clinvar has successfully provided. Clinvar is a public database for clinical laboratories, researchers, expert panels, and others to share their interpretations of variants with their evidence. Gdv showing ‘clinvar variants with precise endpoints’ track next to ncbi human gene annotation. Clinvar recently started accepting submissions that are focused primarily on providing phenotypic information for individuals who have had genetic testing. Aggregating data from many groups in a single database allows comparison of interpretations, providing transparency into the concordance or discordance of interpretations.
 Source: ncbiinsights.ncbi.nlm.nih.gov
Source: ncbiinsights.ncbi.nlm.nih.gov
You have a few options to add the pmid to clinvar •if your vcep will review and submit this variant, include the pmid in your submitted record. Clinvar is a freely available, public archive of human genetic variants and interpretations of their relationships to diseases and other conditions, maintained at the national institutes of health (nih). Variants in clinvar may be of any length or type, ranging from single nucleotide substitutions and small insertions/deletions to copy number changes and cytogenetic rearrangements. Aggregating data from many groups in a single database allows comparison of interpretations, providing transparency into the concordance or discordance of interpretations. • consider batching several citations in one message.

Check through this pubmed listing to locate citations/papers on individual ncbi services of your interest. Check through this pubmed listing to locate citations/papers on individual ncbi services of your interest. Users can leverage this information in their search of the literature for a variant. The reason for the remaining 25% was undetermined because no citation in clinvar or the variant was not found in the citation (column n in the supplementary table s1). Clinvar encourages submissions of variants reviewed by expert panels, as expert consensus confers a high standard.
 Source: researchgate.net
Source: researchgate.net
Submissions may come from clinical providers providing their own interpretation of the variant (�provider interpretation�) or from groups such as patient registries that primarily provide. Citing the ncbi internet site and individual web pages and records: Aggregating data from many groups in a single database allows comparison of interpretations, providing transparency into the concordance or discordance of interpretations. You have a few options to add the pmid to clinvar •if your vcep will review and submit this variant, include the pmid in your submitted record. Gdv showing ‘clinvar variants with precise endpoints’ track next to ncbi human gene annotation.
 Source: researchgate.net
Source: researchgate.net
Clinvar data dictionary, august 4, 2017 page 5 variationrelease xml: Check through this pubmed listing to locate citations/papers on individual ncbi services of your interest. Tracks are color coded for quick and easy interpretation. The subject of this data analysis experiment is a mashup of two datasets, clinvar and the genetic testing registry.please see the linked blog posts for a detailed introduction to these datasets, as well as their technical details and links to formal documentation. Variants in clinvar may be of any length or type, ranging from single nucleotide substitutions and small insertions/deletions to copy number changes and cytogenetic rearrangements.

Of these, 192 (63.4%) were novel to clinvar. There are no citations in clinvar for this variation. For the blast® services, use these blast references. This should be used only when there is no database id for the publication and no url. Clinvar is a public database for clinical laboratories, researchers, expert panels, and others to share their interpretations of variants with their evidence.
 Source: semanticscholar.org
Source: semanticscholar.org
The bottom of the new variation page displays a table of all citations reported to clinvar for the variant with links to pubmed, except for citations provided as assertion criteria and citations that are submitted specifically for the disease (not the variant). Gdv showing ‘clinvar variants with precise endpoints’ track next to ncbi human gene annotation. You have a few options to add the pmid to clinvar •if your vcep will review and submit this variant, include the pmid in your submitted record. Citing the ncbi internet site and individual web pages and records: Clinvar recently started accepting submissions that are focused primarily on providing phenotypic information for individuals who have had genetic testing.
 Source: blog.goldenhelix.com
Source: blog.goldenhelix.com
Users can leverage this information in their search of the literature for a variant. The subject of this data analysis experiment is a mashup of two datasets, clinvar and the genetic testing registry.please see the linked blog posts for a detailed introduction to these datasets, as well as their technical details and links to formal documentation. If you know of citations for this variation, please consider submitting that information to clinvar. Users can leverage this information in their search of the literature for a variant. Of these, 192 (63.4%) were novel to clinvar.

Citing the ncbi internet site and individual web pages and records: •consider a “literature only” submission. You have a few options to add the pmid to clinvar •if your vcep will review and submit this variant, include the pmid in your submitted record. Clinvar data dictionary, august 4, 2017 page 5 variationrelease xml: Clinvar is a public database for clinical laboratories, researchers, expert panels, and others to share their interpretations of variants with their evidence.
This site is an open community for users to submit their favorite wallpapers on the internet, all images or pictures in this website are for personal wallpaper use only, it is stricly prohibited to use this wallpaper for commercial purposes, if you are the author and find this image is shared without your permission, please kindly raise a DMCA report to Us.
If you find this site helpful, please support us by sharing this posts to your preference social media accounts like Facebook, Instagram and so on or you can also save this blog page with the title clinvar citation by using Ctrl + D for devices a laptop with a Windows operating system or Command + D for laptops with an Apple operating system. If you use a smartphone, you can also use the drawer menu of the browser you are using. Whether it’s a Windows, Mac, iOS or Android operating system, you will still be able to bookmark this website.
Category
Related By Category
- Easybib chicago citation information
- Doi to apa citation machine information
- Citation x poh information
- Cpl kyle carpenter medal of honor citation information
- Goethe citation dieu information
- Exact citation apa information
- Citation une impatience information
- Fitzgerald way out there blue citation information
- Contre le racisme citation information
- Friedrich nietzsche citaat grot information