Cuffdiff citation information
Home » Trend » Cuffdiff citation informationYour Cuffdiff citation images are ready. Cuffdiff citation are a topic that is being searched for and liked by netizens today. You can Download the Cuffdiff citation files here. Download all free images.
If you’re searching for cuffdiff citation images information linked to the cuffdiff citation keyword, you have pay a visit to the ideal site. Our site frequently gives you suggestions for downloading the maximum quality video and picture content, please kindly hunt and find more informative video articles and images that match your interests.
Cuffdiff Citation. Please cite this paper if you use cuffdiff in your work. We demonstrate the accuracy of our approach through differential analysis of lung fibroblasts in response to loss of the developmental transcription factor hoxa1, which we show is required for lung fibroblast and hela cell cycle. This issue fixes several bugs: Cufflinks also includes cuffdiff, which accepts the reads assembled from two or more biological conditions and analyzes their differential expression of genes.
 List of differentially expressed genes (from both Cuffdiff From researchgate.net
List of differentially expressed genes (from both Cuffdiff From researchgate.net
Were tabulated using the google scholar citation counts for the respective tool references as of feb. This issue fixes several bugs: However, several package are available for transcript analysis such as rsem… and so one. Cuffdiff 2 detected expression for 16,278 of 69,202 (38%) transcripts in the annotated transcriptome (ucsc hg19 coding genes; Cuffdiff provides analyses of differential expression and regulation at the gene and transcript level. Type �license()� or �licence()� for distribution details.
Moreover, cuffdiff tool is useful for transcript or isoform and differentially splice gene analysis.
The contrast file parser had a problem that could crash cuffdiff. Please cite this paper if you use cuffdiff in your work. Citation counts, percentages of commonly referenced dge tool citations and year of release for edger , cuffdiff/cuffdiff2 [35, 36], deseq2 , limma , degseq , bayseq , samseq , sleuth and noiseq. Cuffdiff/cuffdiff2 [3, 4], deseq2 [6], limma [26], degseq [27], bayseq [28], samseq [29], and noiseq [30]. Were tabulated using the google scholar citation counts for the respective tool references as of feb. Cuffdiff, a part of the cufflinks package, takes the aligned reads from two or more conditions and reports genes and transcripts that are differentially expressed using a.
 Source: researchgate.net
Source: researchgate.net
Cuffdiff/cuffdiff2 [3, 4], deseq2 [6], limma [26], degseq [27], bayseq [28], samseq [29], and noiseq [30]. Type �contributors()� for more information and �citation()� on how to cite r or r packages in publications. Type �license()� or �licence()� for distribution details. Please cite this paper if you use cuffdiff in your work. Cuffnorm was not sometimes permuting replicate numbering, leading to inconsistent expression calls between cuffnorm and cuffdiff.
 Source: researchgate.net
Source: researchgate.net
Comparison of cuffdiff based methods (cuffdiff + subjunc, cuffdiff + subread, cuffdiff + tmap, cuffdiff + tophat2, cuffdiff + tophat2g), rsem and sailfish among a. Cuffdiff calculates the fpkm of each transcript, primary transcript, and gene in each sample. Please cite this paper if you use cuffdiff in your work. R is a collaborative project with many contributors. Several cuffnorm output files had minor output formatting issues.
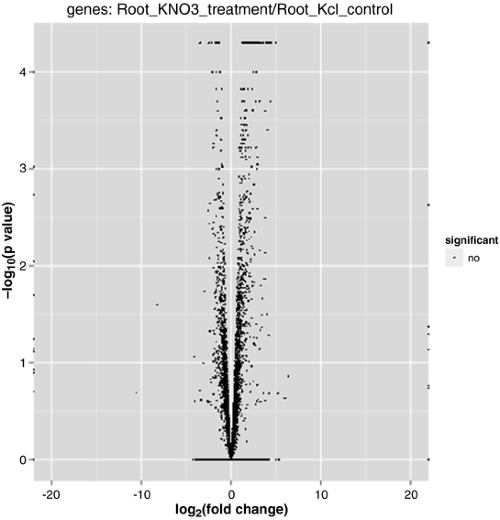 Source: experiments.springernature.com
Source: experiments.springernature.com
Cufflinks builds on many ideas, including some proposed in the following papers: Type �license()� or �licence()� for distribution details. Cufflinks then estimates the relative abundances of these transcripts based on how many reads support. We demonstrate the accuracy of our approach through differential analysis of lung fibroblasts in response to loss of the developmental transcription factor hoxa1, which we show is required for lung fibroblast and hela cell cycle. Cuffdiff provides analyses of differential expression and regulation at the gene and transcript level.
 Source: researchgate.net
Source: researchgate.net
Comparison of cuffdiff based methods (cuffdiff + subjunc, cuffdiff + subread, cuffdiff + tmap, cuffdiff + tophat2, cuffdiff + tophat2g), rsem and sailfish among a. May 05, 2014may 05, 2014permalinkliketweet+1. Cuffdiff calculates the fpkm of each transcript, primary transcript, and gene in each sample. Cuffdiff 2 detected expression for 16,278 of 69,202 (38%) transcripts in the annotated transcriptome (ucsc hg19 coding genes; Citation counts, percentages of commonly referenced dge tool citations and year of release for edger , cuffdiff/cuffdiff2 [35, 36], deseq2 , limma , degseq , bayseq , samseq , sleuth and noiseq.
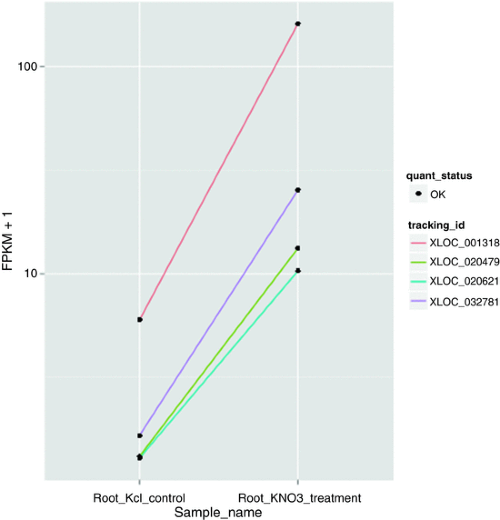 Source: experiments.springernature.com
Source: experiments.springernature.com
Cufflinks also includes cuffdiff, which accepts the reads assembled from two or more biological conditions and analyzes their differential expression of genes. However, several package are available for transcript analysis such as rsem… and so one. Moreover, cuffdiff tool is useful for transcript or isoform and differentially splice gene analysis. Primary transcript and gene fpkms are computed by summing the fpkms of transcripts in each primary transcript group or gene group. Type �contributors()� for more information and �citation()� on how to cite r or r packages in publications.
 Source: researchgate.net
Source: researchgate.net
Cuffdiff calculates the fpkm of each transcript, primary transcript, and gene in each sample. Cuffdiff 2 detected expression for 16,278 of 69,202 (38%) transcripts in the annotated transcriptome (ucsc hg19 coding genes; May 05, 2014may 05, 2014permalinkliketweet+1. Citation counts, percentages of commonly referenced dge tool citations and year of release for edger , cuffdiff/cuffdiff2 [35, 36], deseq2 , limma , degseq , bayseq , samseq , sleuth and noiseq. However, several package are available for transcript analysis such as rsem… and so one.
 Source: researchgate.net
Source: researchgate.net
Cuffdiff provides analyses of differential expression and regulation at the gene and transcript level. Cuffdiff 2 detected expression for 16,278 of 69,202 (38%) transcripts in the annotated transcriptome (ucsc hg19 coding genes; Cufflinks also includes cuffdiff, which accepts the reads assembled from two or more biological conditions and analyzes their differential expression of genes. R is a collaborative project with many contributors. All counts were tabulated using the google scholar citation counts for the respective tool references as of 2 february 2018
 Source: researchgate.net
Source: researchgate.net
Cuffdiff/cuffdiff2 [3, 4], deseq2 [6], limma [26], degseq [27], bayseq [28], samseq [29], and noiseq [30]. Cufflinks also includes cuffdiff, which accepts the reads assembled from two or more biological conditions and analyzes their differential expression of genes. R is a collaborative project with many contributors. May 05, 2014may 05, 2014permalinkliketweet+1. Cuffdiff, a part of the cufflinks package, takes the aligned reads from two or more conditions and reports genes and transcripts that are differentially expressed using a.
 Source: researchgate.net
Source: researchgate.net
May 05, 2014may 05, 2014permalinkliketweet+1. All counts were tabulated using the google scholar citation counts for the respective tool references as of 2 february 2018 R is a collaborative project with many contributors. Moreover, cuffdiff tool is useful for transcript or isoform and differentially splice gene analysis. Were tabulated using the google scholar citation counts for the respective tool references as of feb.
 Source: researchgate.net
Source: researchgate.net
Please cite this paper if you use cuffdiff in your work. You are welcome to redistribute it under certain conditions. The cuffdiff file formats are designed to simplify use by other downstream programs. Cuffnorm was not sometimes permuting replicate numbering, leading to inconsistent expression calls between cuffnorm and cuffdiff. This issue fixes several bugs:
 Source: researchgate.net
Source: researchgate.net
Were tabulated using the google scholar citation counts for the respective tool references as of feb. You are welcome to redistribute it under certain conditions. Cuffdiff/cuffdiff2 [3, 4], deseq2 [6], limma [26], degseq [27], bayseq [28], samseq [29], and noiseq [30]. Cuffdiff provides analyses of differential expression and regulation at the gene and transcript level. Were tabulated using the google scholar citation counts for the respective tool references as of feb.
 Source: researchgate.net
Source: researchgate.net
This issue fixes several bugs: Cuffdiff 2 detected expression for 16,278 of 69,202 (38%) transcripts in the annotated transcriptome (ucsc hg19 coding genes; Citation counts, percentages of commonly referenced dge tool citations and year of release for edger , cuffdiff/cuffdiff2 [35, 36], deseq2 , limma , degseq , bayseq , samseq , sleuth and noiseq. Cuffdiff, a part of the cufflinks package, takes the aligned reads from two or more conditions and reports genes and transcripts that are differentially expressed using a. R is a collaborative project with many contributors.
 Source: researchgate.net
Source: researchgate.net
The cuffdiff file formats are designed to simplify use by other downstream programs. The contrast file parser had a problem that could crash cuffdiff. R is a collaborative project with many contributors. Several cuffnorm output files had minor output formatting issues. May 05, 2014may 05, 2014permalinkliketweet+1.
 Source: researchgate.net
Source: researchgate.net
Comparison of cuffdiff based methods (cuffdiff + subjunc, cuffdiff + subread, cuffdiff + tmap, cuffdiff + tophat2, cuffdiff + tophat2g), rsem and sailfish among a. However, several package are available for transcript analysis such as rsem… and so one. Cuffdiff, a part of the cufflinks package, takes the aligned reads from two or more conditions and reports genes and transcripts that are differentially expressed using a. Cufflinks builds on many ideas, including some proposed in the following papers: Cuffdiff calculates the fpkm of each transcript, primary transcript, and gene in each sample.
 Source: researchgate.net
Source: researchgate.net
All counts were tabulated using the google scholar citation counts for the respective tool references as of 2 february 2018 The contrast file parser had a problem that could crash cuffdiff. Type �license()� or �licence()� for distribution details. Comparison of cuffdiff based methods (cuffdiff + subjunc, cuffdiff + subread, cuffdiff + tmap, cuffdiff + tophat2, cuffdiff + tophat2g), rsem and sailfish among a. Were tabulated using the google scholar citation counts for the respective tool references as of feb.
 Source: researchgate.net
Source: researchgate.net
This file includes all of the results of expression quantification and differential expression detection produced by cuffdiff, following the parameters given in command jobs used to generate differential expression data in experiment 1. You are welcome to redistribute it under certain conditions. The contrast file parser had a problem that could crash cuffdiff. This file includes all of the results of expression quantification and differential expression detection produced by cuffdiff, following the parameters given in command jobs used to generate differential expression data in experiment 1. Type �contributors()� for more information and �citation()� on how to cite r or r packages in publications.
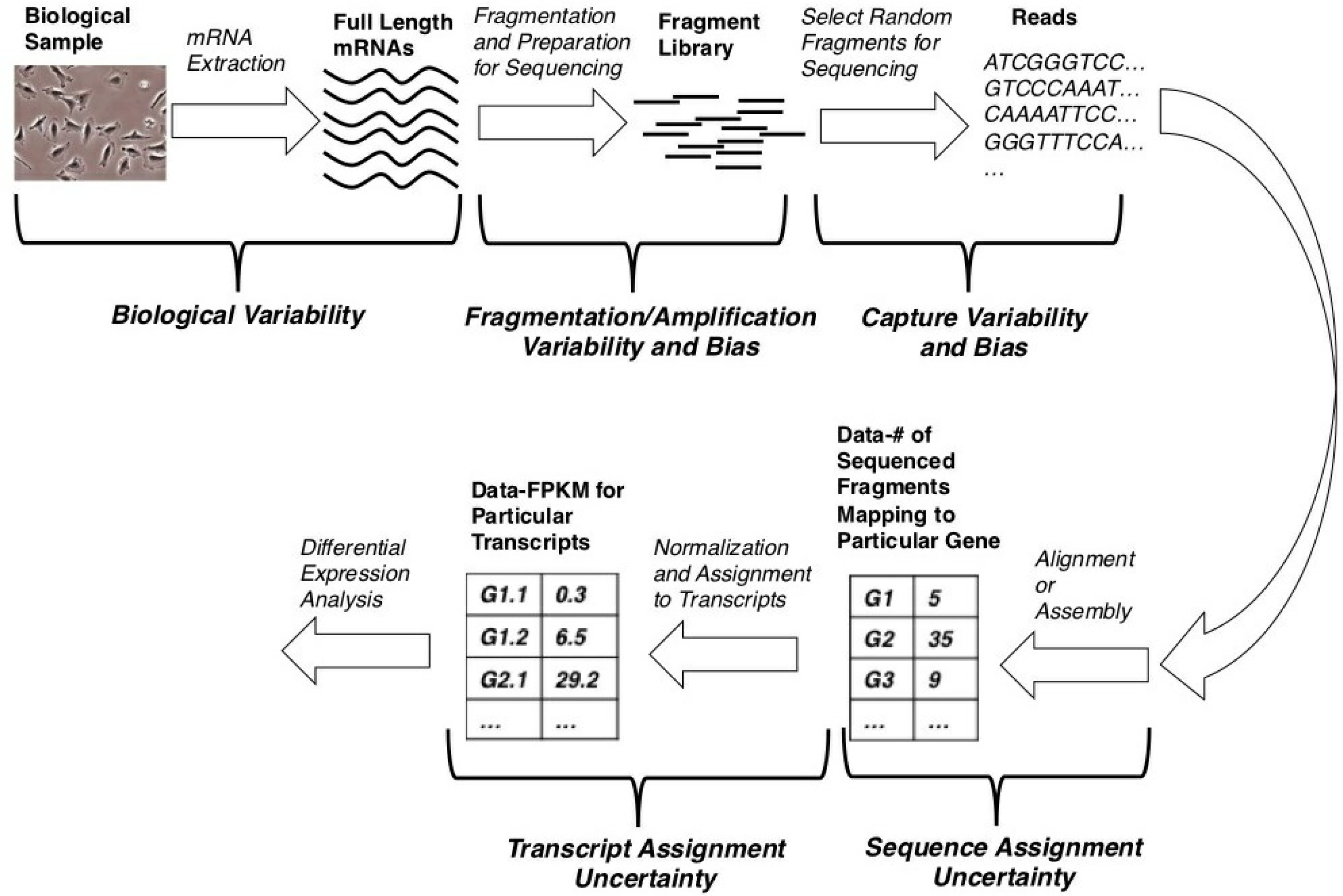 Source: mdpi.com
Source: mdpi.com
All counts were tabulated using the google scholar citation counts for the respective tool references as of 2 february 2018 Citation counts, percentages of commonly referenced dge tool citations and year of release for edger , cuffdiff/cuffdiff2 [35, 36], deseq2 , limma , degseq , bayseq , samseq , sleuth and noiseq. Type �contributors()� for more information and �citation()� on how to cite r or r packages in publications. Alternative isoform abundances relative to one another were maintained in most. The contrast file parser had a problem that could crash cuffdiff.
 Source: researchgate.net
Source: researchgate.net
We demonstrate the accuracy of our approach through differential analysis of lung fibroblasts in response to loss of the developmental transcription factor hoxa1, which we show is required for lung fibroblast and hela cell cycle. May 05, 2014may 05, 2014permalinkliketweet+1. Cufflinks also includes cuffdiff, which accepts the reads assembled from two or more biological conditions and analyzes their differential expression of genes. Type �contributors()� for more information and �citation()� on how to cite r or r packages in publications. Citation counts, percentages of commonly referenced dge tool citations and year of release for edger , cuffdiff/cuffdiff2 [35, 36], deseq2 , limma , degseq , bayseq , samseq , sleuth and noiseq.
This site is an open community for users to do submittion their favorite wallpapers on the internet, all images or pictures in this website are for personal wallpaper use only, it is stricly prohibited to use this wallpaper for commercial purposes, if you are the author and find this image is shared without your permission, please kindly raise a DMCA report to Us.
If you find this site helpful, please support us by sharing this posts to your preference social media accounts like Facebook, Instagram and so on or you can also bookmark this blog page with the title cuffdiff citation by using Ctrl + D for devices a laptop with a Windows operating system or Command + D for laptops with an Apple operating system. If you use a smartphone, you can also use the drawer menu of the browser you are using. Whether it’s a Windows, Mac, iOS or Android operating system, you will still be able to bookmark this website.
Category
Related By Category
- De vliegeraar citaten information
- Full reference citation apa style information
- Free apa citation machine online information
- Etre amoureux citation information
- Fight club citation tyler information
- Evene lefigaro fr citations information
- Freud citations aimer et travailler information
- Endnote book citation information
- Flap lever cessna citation information
- Foreign aid debate citation information