Cutadapt citation information
Home » Trend » Cutadapt citation informationYour Cutadapt citation images are ready. Cutadapt citation are a topic that is being searched for and liked by netizens today. You can Get the Cutadapt citation files here. Get all free photos.
If you’re searching for cutadapt citation pictures information related to the cutadapt citation keyword, you have come to the right site. Our site frequently provides you with hints for seeing the maximum quality video and picture content, please kindly surf and locate more informative video articles and graphics that match your interests.
Cutadapt Citation. As a result, we recommend using the 2020 book as a citation over the 2013 paper. A tool that removes adapter sequences from dna sequencing reads. When forward and reverse reads coexist in. , 2014 ), another popular trimming adapter tool, can perform quality pruning using algorithms such as sliding window cutting.
 Quality control statistical distribution of Cutadapt From researchgate.net
Quality control statistical distribution of Cutadapt From researchgate.net
Cutadapt searches for the adapter. As a result, we recommend using the 2020 book as a citation over the 2013 paper. You should see the version number of cutadapt. A functional version of cutadapt and optionally fastqc are required. Cleaning your data in this way is often required: However, by comparing data analysis results of preprocessing with cutadapt, fastp, trimmomatic, and raw.
, 2014 ), another popular trimming adapter tool, can perform quality pruning using algorithms such as sliding window cutting.
It is currently being developed within nbis (national bioinformatics infrastructure sweden). A functional version of cutadapt and optionally fastqc are required. If you use cutadapt, please cite doi:10.14806/ej. Van der auwera ga, carneiro m, hartl c, poplin r. Note, the cutadapt default of 0 has been overridden, because that value produces empty sequence records. We would like to show you a description here but the site won’t allow us.
 Source: researchgate.net
Source: researchgate.net
A tool that removes adapter sequences from dna sequencing reads. , 2014 ), another popular trimming adapter tool, can perform quality pruning using algorithms such as sliding window cutting. If you use cutadapt, please cite doi:10.14806/ej. As a result, we recommend using the 2020 book as a citation over the 2013 paper. However, by comparing data analysis results of preprocessing with cutadapt, fastp, trimmomatic, and raw.
 Source: researchgate.net
Source: researchgate.net
It is currently being developed within nbis (national bioinformatics infrastructure sweden). Trimmomatic ( bolger et al. We would like to show you a description here but the site won’t allow us. However, by comparing data analysis results of preprocessing with cutadapt, fastp, trimmomatic, and raw. If you use cutadapt, please cite doi:10.14806/ej.
 Source: researchgate.net
Source: researchgate.net
We would like to show you a description here but the site won’t allow us. Cleaning your data in this way is often required: A tool that removes adapter sequences from dna sequencing reads. Has been cited by the following article: We would like to show you a description here but the site won’t allow us.
 Source: researchgate.net
Source: researchgate.net
Note, the cutadapt default of 0 has been overridden, because that value produces empty sequence records. Trimmomatic ( bolger et al. Cutadapt searches for the adapter. As a result, we recommend using the 2020 book as a citation over the 2013 paper. If you use cutadapt, please cite doi:10.14806/ej.
 Source: researchgate.net
Source: researchgate.net
Using docker, gatk, and wdl in terra. It is currently being developed within nbis (national bioinformatics infrastructure sweden). When small rna is sequenced on current sequencing machines, the resulting reads are usually longer than the rna and therefore contain parts of the 3� adapter. Cutadapt supports parallel processing, that is, it can use multiple cpu cores. However, by comparing data analysis results of preprocessing with cutadapt, fastp, trimmomatic, and raw.
![[NGS QC] MultiQC BioinformaticsAndMe [NGS QC] MultiQC BioinformaticsAndMe](https://img1.daumcdn.net/thumb/R800x0/?scode=mtistory2&fname=https:%2F%2Ft1.daumcdn.net%2Fcfile%2Ftistory%2F99E18B3A5B6FBD331D)
The detection takes into account resource restrictions that may be in place. When forward and reverse reads coexist in. Using docker, gatk, and wdl in terra. , 2014 ), another popular trimming adapter tool, can perform quality pruning using algorithms such as sliding window cutting. Van der auwera ga, carneiro m, hartl c, poplin r.
 Source: nekrut.github.io
Source: nekrut.github.io
As a result, we recommend using the 2020 book as a citation over the 2013 paper. However, by comparing data analysis results of preprocessing with cutadapt, fastp, trimmomatic, and raw. The detection takes into account resource restrictions that may be in place. If you use cutadapt, please cite doi:10.14806/ej. Using docker, gatk, and wdl in terra.

The detection takes into account resource restrictions that may be in place. However, by comparing data analysis results of preprocessing with cutadapt, fastp, trimmomatic, and raw. Using docker, gatk, and wdl in terra. Has been cited by the following article: Note, the cutadapt default of 0 has been overridden, because that value produces empty sequence records.
 Source: researchgate.net
Source: researchgate.net
Cutadapt searches for the adapter. Trimmomatic ( bolger et al. Has been cited by the following article: Cleaning your data in this way is often required: A tool that removes adapter sequences from dna sequencing reads.
 Source: researchgate.net
Source: researchgate.net
Note, the cutadapt default of 0 has been overridden, because that value produces empty sequence records. Cleaning your data in this way is often required: The detection takes into account resource restrictions that may be in place. You should see the version number of cutadapt. Note, the cutadapt default of 0 has been overridden, because that value produces empty sequence records.
 Source: researchgate.net
Source: researchgate.net
Trimmomatic ( bolger et al. Note, the cutadapt default of 0 has been overridden, because that value produces empty sequence records. When forward and reverse reads coexist in. You should see the version number of cutadapt. When small rna is sequenced on current sequencing machines, the resulting reads are usually longer than the rna and therefore contain parts of the 3� adapter.
 Source: researchgate.net
Source: researchgate.net
As a result, we recommend using the 2020 book as a citation over the 2013 paper. Cleaning your data in this way is often required: When forward and reverse reads coexist in. We would like to show you a description here but the site won’t allow us. Cutadapt supports parallel processing, that is, it can use multiple cpu cores.
 Source: researchgate.net
Source: researchgate.net
As a result, we recommend using the 2020 book as a citation over the 2013 paper. We would like to show you a description here but the site won’t allow us. Note, the cutadapt default of 0 has been overridden, because that value produces empty sequence records. , 2014 ), another popular trimming adapter tool, can perform quality pruning using algorithms such as sliding window cutting. Cutadapt searches for the adapter.
Source: researchgate.net
However, by comparing data analysis results of preprocessing with cutadapt, fastp, trimmomatic, and raw. However, by comparing data analysis results of preprocessing with cutadapt, fastp, trimmomatic, and raw. Note, the cutadapt default of 0 has been overridden, because that value produces empty sequence records. When small rna is sequenced on current sequencing machines, the resulting reads are usually longer than the rna and therefore contain parts of the 3� adapter. A tool that removes adapter sequences from dna sequencing reads.
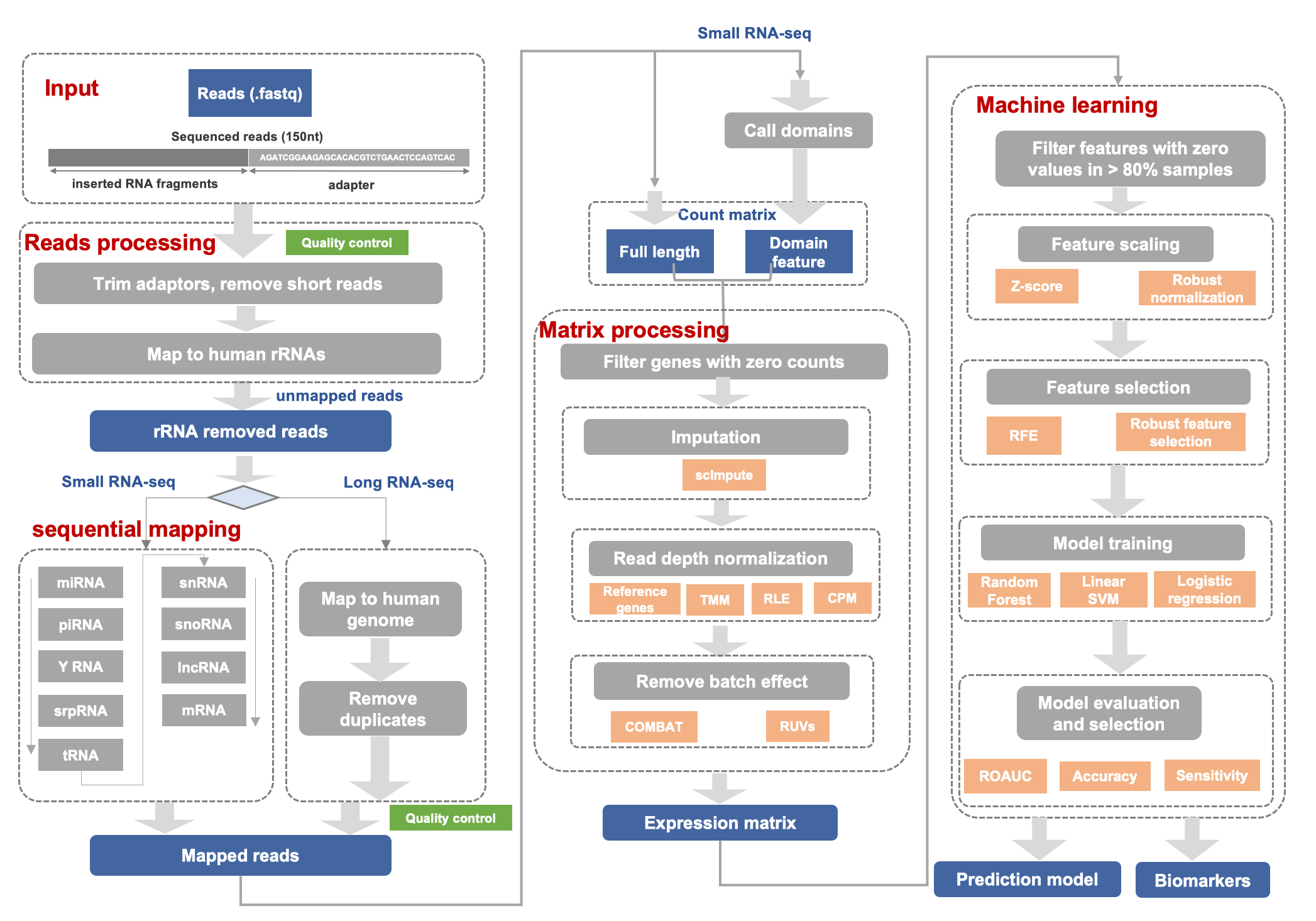 Source: lulab.github.io
Source: lulab.github.io
Cleaning your data in this way is often required: When forward and reverse reads coexist in. As a result, we recommend using the 2020 book as a citation over the 2013 paper. Note, the cutadapt default of 0 has been overridden, because that value produces empty sequence records. It is currently being developed within nbis (national bioinformatics infrastructure sweden).
 Source: researchgate.net
Source: researchgate.net
As a result, we recommend using the 2020 book as a citation over the 2013 paper. The detection takes into account resource restrictions that may be in place. Using docker, gatk, and wdl in terra. Cutadapt searches for the adapter. You should see the version number of cutadapt.
 Source: researchgate.net
Source: researchgate.net
Has been cited by the following article: If you use cutadapt, please cite doi:10.14806/ej. As a result, we recommend using the 2020 book as a citation over the 2013 paper. It is currently being developed within nbis (national bioinformatics infrastructure sweden). The detection takes into account resource restrictions that may be in place.
 Source: researchgate.net
Source: researchgate.net
A tool that removes adapter sequences from dna sequencing reads. Van der auwera ga, carneiro m, hartl c, poplin r. When forward and reverse reads coexist in. Cutadapt supports parallel processing, that is, it can use multiple cpu cores. Has been cited by the following article:
This site is an open community for users to share their favorite wallpapers on the internet, all images or pictures in this website are for personal wallpaper use only, it is stricly prohibited to use this wallpaper for commercial purposes, if you are the author and find this image is shared without your permission, please kindly raise a DMCA report to Us.
If you find this site value, please support us by sharing this posts to your preference social media accounts like Facebook, Instagram and so on or you can also save this blog page with the title cutadapt citation by using Ctrl + D for devices a laptop with a Windows operating system or Command + D for laptops with an Apple operating system. If you use a smartphone, you can also use the drawer menu of the browser you are using. Whether it’s a Windows, Mac, iOS or Android operating system, you will still be able to bookmark this website.
Category
Related By Category
- De vliegeraar citaten information
- Full reference citation apa style information
- Free apa citation machine online information
- Etre amoureux citation information
- Fight club citation tyler information
- Evene lefigaro fr citations information
- Freud citations aimer et travailler information
- Endnote book citation information
- Flap lever cessna citation information
- Foreign aid debate citation information