Dbsnp citation information
Home » Trend » Dbsnp citation informationYour Dbsnp citation images are available in this site. Dbsnp citation are a topic that is being searched for and liked by netizens now. You can Find and Download the Dbsnp citation files here. Download all free photos.
If you’re searching for dbsnp citation pictures information related to the dbsnp citation keyword, you have pay a visit to the right site. Our website always gives you suggestions for seeing the highest quality video and picture content, please kindly search and locate more enlightening video articles and images that match your interests.
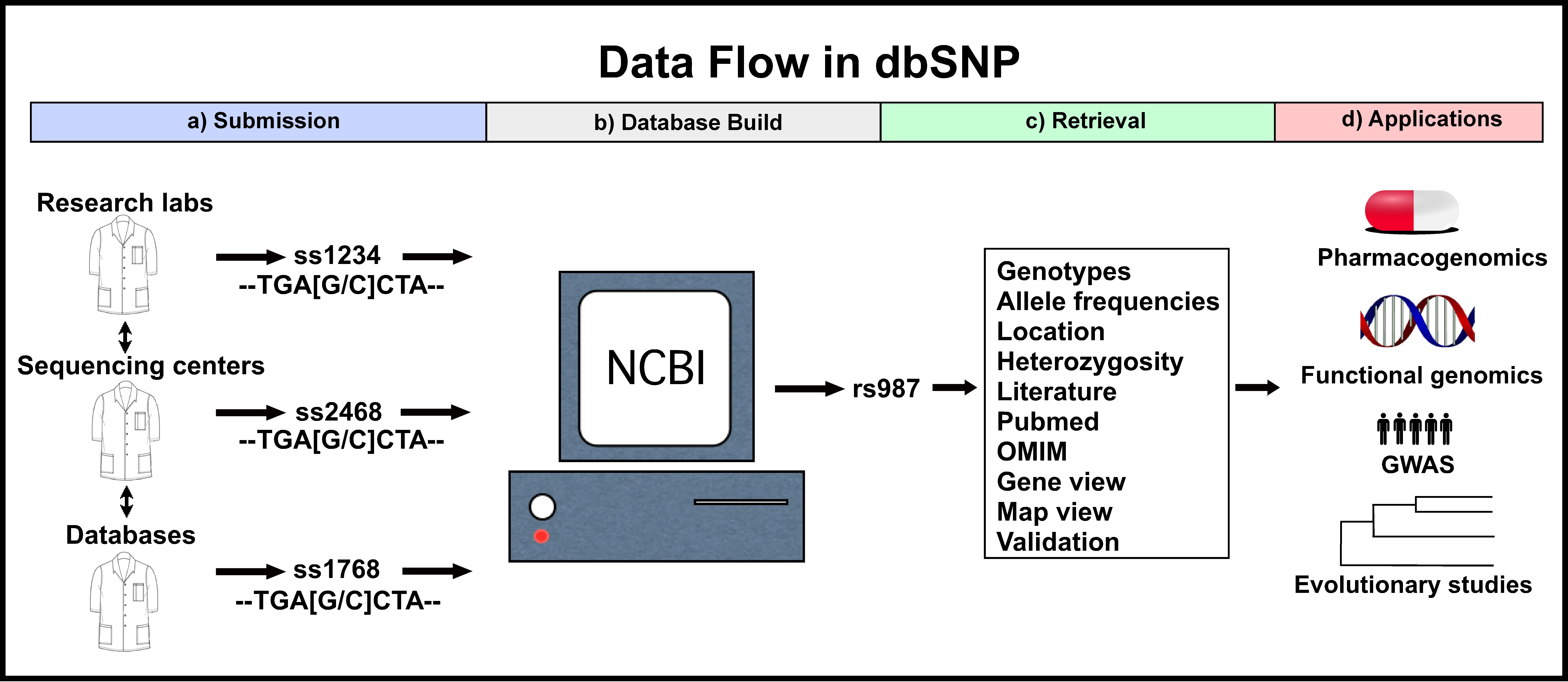
Dbsnp Citation. The variant is reported to be polymorphic in at least one sample. The variant has multiple independent dbsnp submissions, i.e. A surveillance system that monitors data repositories and reports changes helps manage the data overload. Submissions with a different submitter handles or different discovery samples.
 Example results page from the NCBI dbSNP database for From researchgate.net
Example results page from the NCBI dbSNP database for From researchgate.net
Annotating id from dbsnp $ cat test.chr22.vcf #chrom pos id ref alt qual filter info 22 16157571. The source data files used for this package were created by ncbi on 9 november 2010 and contain snps mapped to reference genome grch37. Dbsnp was established in august 1999 as a collaboration between ncbi and the national human genome research institute (nhgri) as a database of small scale nucleotide variants. Snp locations and alleles for homo sapiens extracted from dbsnp build 131:human_9606 (based on grch37). Dbsnp was established in august 1999 as a collaboration between ncbi and the national human genome research institute (nhgri) as a database of small scale nucleotide variants. However, users can also retrieve older versions of dbsnp:
Dbsnp was established in august 1999 as a collaboration between ncbi and the national human genome research institute (nhgri) as a database of small scale nucleotide variants.
(1) convert genome position from one genome assembly to another genome assembly in most scenarios, we have known genome positions in ncbi build 36 (ucsc hg 18) and hope to lift them over to ncbi build 37 (ucsc hg19). Snp locations and alleles for homo sapiens extracted from dbsnp build 131:human_9606 (based on grch37). The snps in this package are based on the grch37 genome. Submissions with a different submitter handles or different discovery samples. Snp locations and alleles for homo sapiens extracted from ncbi dbsnp build 137. However, users can also retrieve older versions of dbsnp:
 Source: researchgate.net
Source: researchgate.net
Annotating id from dbsnp $ cat test.chr22.vcf #chrom pos id ref alt qual filter info 22 16157571. Dbsnp was established in august 1999 as a collaboration between ncbi and the national human genome research institute (nhgri) as a database of small scale nucleotide variants. Liftover is a necesary step to bring all genetical analysis to the same reference build. Dbsnp is a free public archive for genetic variation within and across different species developed and hosted by the national center for biotechnology information (ncbi) in collaboration with the national human genome research institute (nhgri). Snp locations and alleles for homo sapiens extracted from ncbi dbsnp build 132.
 Source: researchgate.net
Source: researchgate.net
The technical redesign prepares the database for increasing data volumes and providing timely, effective and trustworthy reference snp results as submission rates continue to increase. The default version of our dbsnp annotation is currently referring to dbsnp143 (using hg38 coordinates) as shown below. Dbsnp was established in august 1999 as a collaboration between ncbi and the national human genome research institute (nhgri) as a database of small scale nucleotide variants. Snp locations and alleles for homo sapiens extracted from ncbi dbsnp build 137. The variant is reported to be polymorphic in at least one sample.
 Source: researchgate.net
Source: researchgate.net
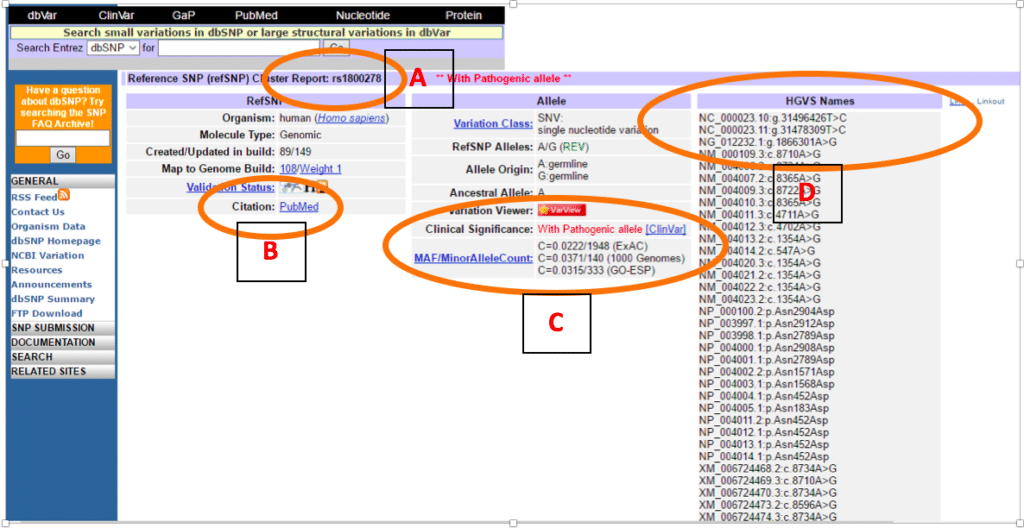
T g 0.0 fail ns=53 22 16346045. The default version of our dbsnp annotation is currently referring to dbsnp143 (using hg38 coordinates) as shown below. Dbsnp web query for build 155: T g 0.0 fail ns=53 22 16346045. The variant is cited in a pubmed article.
 Source: researchgate.net
Source: researchgate.net
Links to the literature databases are made with the citation information provided at submission time. Dbsnp currently classifies n ucleotide sequence v ariations with. Dbsnp is a free public archive for genetic variation within and across different species developed and hosted by the national center for biotechnology information (ncbi) in collaboration with the national human genome research institute (nhgri). Liftover can have three use cases: The default version of our dbsnp annotation is currently referring to dbsnp143 (using hg38 coordinates) as shown below.
 Source: researchgate.net
Source: researchgate.net
Dbsnp currently classifies n ucleotide sequence v ariations with. Dbsnp was established in august 1999 as a collaboration between ncbi and the national human genome research institute (nhgri) as a database of small scale nucleotide variants. It is a public repository of submitted nucleotide variations and is part of ncbi�s search and retrieval system entrez. The variant has multiple independent dbsnp submissions, i.e. Liftover is a necesary step to bring all genetical analysis to the same reference build.
 Source: researchgate.net
Source: researchgate.net
Liftover is a necesary step to bring all genetical analysis to the same reference build. The snps in this package are based on the grch37 genome. Note that the grch37 genome is the same as the hg19 genome from ucsc except for the mitochondrion chromosome. Liftover can have three use cases: The variant has multiple independent dbsnp submissions, i.e.
 Source: writework.com
Source: writework.com
The variant has multiple independent dbsnp submissions, i.e. The variant is cited in a pubmed article. Use of molecular variation in the ncbi. Dbsnp was established in august 1999 as a collaboration between ncbi and the national human genome research institute (nhgri) as a database of small scale nucleotide variants. The 129 and 130 versions use hg18 as a reference genome, 131, 132, 135, 137, 138 and 141.
 Source: researchgate.net
Source: researchgate.net
The following types a nd percentage composition of the database: Snp locations and alleles for homo sapiens extracted from dbsnp build 131:human_9606 (based on grch37). Dbsnp was established in august 1999 as a collaboration between ncbi and the national human genome research institute (nhgri) as a database of small scale nucleotide variants. (1) convert genome position from one genome assembly to another genome assembly in most scenarios, we have known genome positions in ncbi build 36 (ucsc hg 18) and hope to lift them over to ncbi build 37 (ucsc hg19). Dbsnp141, dbsnp138, dbsnp137, dbsnp135, dbsnp132, dbsnp131, dbsnp130, dbsnp129.
 Source: researchgate.net
Source: researchgate.net
Note that the grch37 genome is the same as the hg19 genome from ucsc except. Submissions with a different submitter handles or different discovery samples. Entrez indexing for build 155: To continue providing efficient and timely processing, annotation, and dissemination of data, dbsnp’s architecture and process flow have been redesigned. Dbsnp141, dbsnp138, dbsnp137, dbsnp135, dbsnp132, dbsnp131, dbsnp130, dbsnp129.
 Source: researchgate.net
Source: researchgate.net
(1) convert genome position from one genome assembly to another genome assembly in most scenarios, we have known genome positions in ncbi build 36 (ucsc hg 18) and hope to lift them over to ncbi build 37 (ucsc hg19). Liftover is a necesary step to bring all genetical analysis to the same reference build. Ftp data for build 155: Note that the grch37 genome is the same as the hg19 genome from ucsc except for the mitochondrion chromosome. The following types a nd percentage composition of the database:
 Source: researchgate.net
Source: researchgate.net
It is a public repository of submitted nucleotide variations and is part of ncbi�s search and retrieval system entrez. Liftover can have three use cases: Snp locations and alleles for homo sapiens extracted from dbsnp build 131:human_9606 (based on grch37). The default version of our dbsnp annotation is currently referring to dbsnp143 (using hg38 coordinates) as shown below. The source data files used for this package were created by ncbi on 9 november 2010 and contain snps mapped to reference genome grch37.
 Source: researchgate.net
Source: researchgate.net
Annotating id from dbsnp $ cat test.chr22.vcf #chrom pos id ref alt qual filter info 22 16157571. Entrez indexing for build 155: Submissions with a different submitter handles or different discovery samples. However, users can also retrieve older versions of dbsnp: The variant has multiple independent dbsnp submissions, i.e.
 Source: researchgate.net
Source: researchgate.net
The variant has multiple independent dbsnp submissions, i.e. The default version of our dbsnp annotation is currently referring to dbsnp143 (using hg38 coordinates) as shown below. To continue providing efficient and timely processing, annotation, and dissemination of data, dbsnp’s architecture and process flow have been redesigned. Note that the grch37 genome is the same as the hg19 genome from ucsc except. Ftp data for build 155:
 Source: researchgate.net
Source: researchgate.net
The variant is cited in a pubmed article. Snp locations and alleles for homo sapiens extracted from ncbi dbsnp build 132. Entrez indexing for build 155: Note that the grch37 genome is the same as the hg19 genome from ucsc except. Note that the grch37 genome is the same as the hg19 genome from ucsc except for the mitochondrion chromosome.
 Source: researchgate.net
Source: researchgate.net
Submissions with a different submitter handles or different discovery samples. Annotating id from dbsnp $ cat test.chr22.vcf #chrom pos id ref alt qual filter info 22 16157571. Dbsnp is moving to the new design with new products ready for testing including new json data files, the refsnp page, and an api. A surveillance system that monitors data repositories and reports changes helps manage the data overload. Snp locations and alleles for homo sapiens extracted from dbsnp build 131:human_9606 (based on grch37).
 Source: bitesizebio.com
Source: bitesizebio.com
The following types a nd percentage composition of the database: Dbsnp was established in august 1999 as a collaboration between ncbi and the national human genome research institute (nhgri) as a database of small scale nucleotide variants. Snp locations and alleles for homo sapiens extracted from dbsnp build 131:human_9606 (based on grch37). To continue providing efficient and timely processing, annotation, and dissemination of data, dbsnp’s architecture and process flow have been redesigned. The variant is cited in a pubmed article.
 Source: researchgate.net
Source: researchgate.net
Liftover can have three use cases: Note that the grch37 genome is the same as the hg19 genome from ucsc except for the mitochondrion chromosome. Liftover can have three use cases: Dbsnp is a free public archive for genetic variation within and across different species developed and hosted by the national center for biotechnology information (ncbi) in collaboration with the national human genome research institute (nhgri). A surveillance system that monitors data repositories and reports changes helps manage the data overload.
 Source: researchgate.net
Source: researchgate.net
Links to the literature databases are made with the citation information provided at submission time. A surveillance system that monitors data repositories and reports changes helps manage the data overload. However, users can also retrieve older versions of dbsnp: Liftover can have three use cases: T g 0.0 fail ns=53 22 16346045.
This site is an open community for users to do sharing their favorite wallpapers on the internet, all images or pictures in this website are for personal wallpaper use only, it is stricly prohibited to use this wallpaper for commercial purposes, if you are the author and find this image is shared without your permission, please kindly raise a DMCA report to Us.
If you find this site good, please support us by sharing this posts to your favorite social media accounts like Facebook, Instagram and so on or you can also bookmark this blog page with the title dbsnp citation by using Ctrl + D for devices a laptop with a Windows operating system or Command + D for laptops with an Apple operating system. If you use a smartphone, you can also use the drawer menu of the browser you are using. Whether it’s a Windows, Mac, iOS or Android operating system, you will still be able to bookmark this website.
Category
Related By Category
- De vliegeraar citaten information
- Full reference citation apa style information
- Free apa citation machine online information
- Etre amoureux citation information
- Fight club citation tyler information
- Evene lefigaro fr citations information
- Freud citations aimer et travailler information
- Endnote book citation information
- Flap lever cessna citation information
- Foreign aid debate citation information