Gene ontology citation information
Home » Trend » Gene ontology citation informationYour Gene ontology citation images are available. Gene ontology citation are a topic that is being searched for and liked by netizens now. You can Find and Download the Gene ontology citation files here. Get all royalty-free images.
If you’re looking for gene ontology citation pictures information connected with to the gene ontology citation keyword, you have come to the ideal blog. Our website frequently provides you with hints for seeing the highest quality video and picture content, please kindly surf and locate more informative video content and images that match your interests.
Gene Ontology Citation. The gene ontology annotation (goa) database. The gene association files ingested from go consortium members are shown in the table below. Human genes belonging to gene ontology (go) molecular function and biological processes terms were retrieved by unifying iteratively all genes belonging to all child terms of a. In general, an ontology such as the gene ontology consists of a number of explicitly defined terms that are names for biological objects or events.
 Gene Ontology enrichment for differentially expressed From researchgate.net
Gene Ontology enrichment for differentially expressed From researchgate.net
Ashburner, m., ball, c., blake, j. Human genes belonging to gene ontology (go) molecular function and biological processes terms were retrieved by unifying iteratively all genes belonging to all child terms of a. For each citation the type of evidence will be encoded. Each term describes details about this gene’s molecular function,. These terms are depicted as nodes (also called vertices) in a graph that describe the relationships between the nodes. For example, the homeobox d9a gene product from zebrafish has numerous go terms associated with it.
See the go citation guide for citing the go project.
Mouse genome database (mgd) 2019. These terms are depicted as nodes (also called vertices) in a graph that describe the relationships between the nodes. Bult cj, blake ja, smith cl, kadin ja, richardson je, the mouse genome database group. The gene ontology annotation (goa) database. Human genes belonging to gene ontology (go) molecular function and biological processes terms were retrieved by unifying iteratively all genes belonging to all child terms of a. Header describing the date generated and the go release used, for example:
 Source: researchgate.net
Source: researchgate.net
The gene ontology (go) knowledgebase is the world’s largest source of information on the functions of genes. The gene ontology allows users to describe a gene/gene product in detail, considering three main aspects: Bult cj, blake ja, smith cl, kadin ja, richardson je, the mouse genome database group. The gene ontology annotation (goa) database. Tool for the unification of biology.
 Source: researchgate.net
Source: researchgate.net
In general, an ontology such as the gene ontology consists of a number of explicitly defined terms that are names for biological objects or events. Human genes belonging to gene ontology (go) molecular function and biological processes terms were retrieved by unifying iteratively all genes belonging to all child terms of a. As of early april 2000 there were 1,923, 2,094 and 490 nodes in the process, function and component ontologies, respectively. Introduction gene ontology is developing ontologies and frameworks to help annotate biology in a consistent way and help users to integrate and reuse our data in their approaches. The gene association files ingested from go consortium members are shown in the table below.
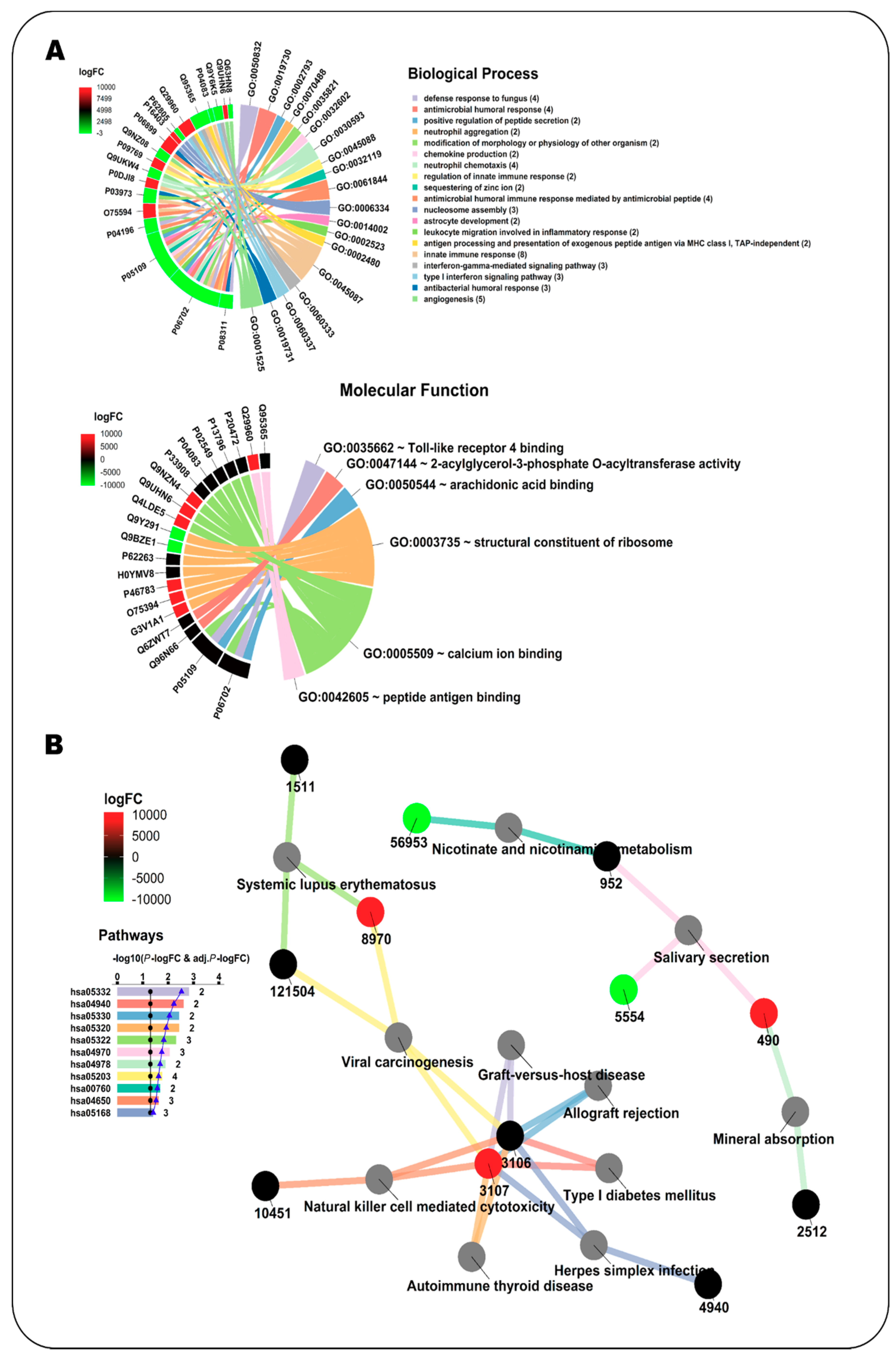 Source: mdpi.com
Source: mdpi.com
In general, an ontology such as the gene ontology consists of a number of explicitly defined terms that are names for biological objects or events. Each term describes details about this gene’s molecular function,. The gene association files ingested from go consortium members are shown in the table below. Header describing the date generated and the go release used, for example: Please use the following citation formats when referring to resources available from the mouse genome informatics web site in your publications.
 Source: researchgate.net
Source: researchgate.net
Tool for the unification of biology. Gaudet p., livstone m.s., lewis s.e., thomas p.d. Tool for the unification of biology. Researchers who notice either missing or inaccurate annotations for a gene or a go term can send this information to the go consortium. Any errors or omissions in annotations should be reported by writing.
 Source: researchgate.net
Source: researchgate.net
The gene ontology (go) project is a major bioinformatics initiative to develop a computational representation of our evolving knowledge of how genes encode biological functions at the molecular, cellular and tissue system levels. Researchers who notice either missing or inaccurate annotations for a gene or a go term can send this information to the go consortium. The gene ontology (go) project is a major bioinformatics initiative to develop a computational representation of our evolving knowledge of how genes encode biological functions at the molecular, cellular and tissue system levels. Files are in the go annotation file format and are compressed using the unix gzip utility. Mouse genome database (mgd) 2019.
 Source: researchgate.net
Source: researchgate.net
The gene ontology annotation (goa) database. Each term describes details about this gene’s molecular function,. Header describing the date generated and the go release used, for example: 2000) provides a hierarchical vocabulary of go terms for describing functions and characteristics of gene or gene products in different databases and different species. The gene ontology allows users to describe a gene/gene product in detail, considering three main aspects:
 Source: researchgate.net
Source: researchgate.net
Introduction gene ontology is developing ontologies and frameworks to help annotate biology in a consistent way and help users to integrate and reuse our data in their approaches. As of early april 2000 there were 1,923, 2,094 and 490 nodes in the process, function and component ontologies, respectively. Gaudet p., livstone m.s., lewis s.e., thomas p.d. The gene ontology (go) knowledgebase is the world’s largest source of information on the functions of genes. In general, an ontology such as the gene ontology consists of a number of explicitly defined terms that are names for biological objects or events.
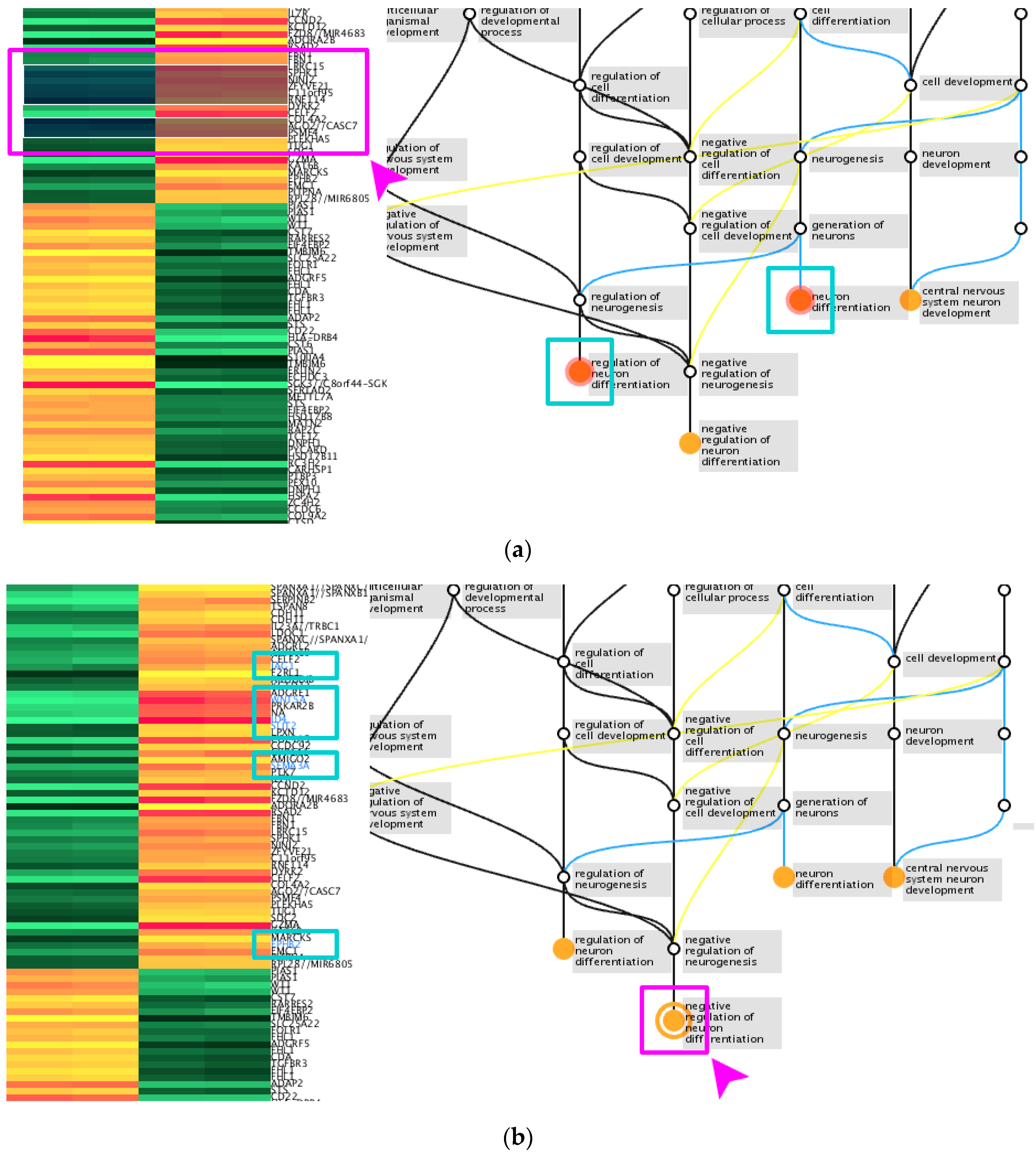 Source: mdpi.com
Source: mdpi.com
The gene ontology (go) knowledgebase is the world’s largest source of information on the functions of genes. The gene product and the term combination that you suggest be reviewed and why you believe this is incorrect Files are in the go annotation file format and are compressed using the unix gzip utility. The gene ontology (go) project is a major bioinformatics initiative to develop a computational representation of our evolving knowledge of how genes encode biological functions at the molecular, cellular and tissue system levels. Header describing the date generated and the go release used, for example:
 Source: researchgate.net
Source: researchgate.net
Researchers who notice either missing or inaccurate annotations for a gene or a go term can send this information to the go consortium. Any errors or omissions in annotations should be reported by writing. The goal of the gene ontology (go) project is to provide a uniform way to describe the functions of gene products from organisms across all kingdoms of life and thereby enable analysis of genomic data. Bingo can be used either on a list of genes, pasted as text, or interactively on subgraphs of biological networks visualized in cytoscape. Paper citation (pmid if available) description of the issue:
Source: researchgate.net
As of early april 2000 there were 1,923, 2,094 and 490 nodes in the process, function and component ontologies, respectively. Gene ontology (go) (ashburner et al. Paper citation (pmid if available) description of the issue: Ashburner, m., ball, c., blake, j. Mouse genome database (mgd) 2019.
 Source: researchgate.net
Source: researchgate.net
See the go citation guide for citing the go project. Tool for the unification of biology. Bingo can be used either on a list of genes, pasted as text, or interactively on subgraphs of biological networks visualized in cytoscape. Tool for the unification of biology. Header describing the date generated and the go release used, for example:
 Source: researchgate.net
Source: researchgate.net
The goal of the gene ontology (go) project is to provide a uniform way to describe the functions of gene products from organisms across all kingdoms of life and thereby enable analysis of genomic data. Paper citation (pmid if available) description of the issue: Mouse genome database (mgd) 2019. For each citation the type of evidence will be encoded. Tool for the unification of biology.
 Source: researchgate.net
Source: researchgate.net
Introduction gene ontology is developing ontologies and frameworks to help annotate biology in a consistent way and help users to integrate and reuse our data in their approaches. The goal of the gene ontology (go) project is to provide a uniform way to describe the functions of gene products from organisms across all kingdoms of life and thereby enable analysis of genomic data. In general, an ontology such as the gene ontology consists of a number of explicitly defined terms that are names for biological objects or events. Bult cj, blake ja, smith cl, kadin ja, richardson je, the mouse genome database group. Gene ontology (go) (ashburner et al.
 Source: researchgate.net
Source: researchgate.net
See the go citation guide for citing the go project. The gene ontology annotation (goa) database. Please use the following citation formats when referring to resources available from the mouse genome informatics web site in your publications. The gene association files ingested from go consortium members are shown in the table below. Tool for the unification of biology.
 Source: researchgate.net
Source: researchgate.net
See the go citation guide for citing the go project. As of early april 2000 there were 1,923, 2,094 and 490 nodes in the process, function and component ontologies, respectively. Bingo can be used either on a list of genes, pasted as text, or interactively on subgraphs of biological networks visualized in cytoscape. Header describing the date generated and the go release used, for example: Human genes belonging to gene ontology (go) molecular function and biological processes terms were retrieved by unifying iteratively all genes belonging to all child terms of a.
 Source: researchgate.net
Source: researchgate.net
Human genes belonging to gene ontology (go) molecular function and biological processes terms were retrieved by unifying iteratively all genes belonging to all child terms of a. Each term describes details about this gene’s molecular function,. Ashburner, m., ball, c., blake, j. Introduction gene ontology is developing ontologies and frameworks to help annotate biology in a consistent way and help users to integrate and reuse our data in their approaches. The gene ontology (go) knowledgebase is the world’s largest source of information on the functions of genes.
Source: researchgate.net
Human genes belonging to gene ontology (go) molecular function and biological processes terms were retrieved by unifying iteratively all genes belonging to all child terms of a. For example, the homeobox d9a gene product from zebrafish has numerous go terms associated with it. Each term describes details about this gene’s molecular function,. These terms are depicted as nodes (also called vertices) in a graph that describe the relationships between the nodes. Bult cj, blake ja, smith cl, kadin ja, richardson je, the mouse genome database group.
 Source: researchgate.net
Source: researchgate.net
The goal of the gene ontology (go) project is to provide a uniform way to describe the functions of gene products from organisms across all kingdoms of life and thereby enable analysis of genomic data. The gene product and the term combination that you suggest be reviewed and why you believe this is incorrect Any errors or omissions in annotations should be reported by writing. Header describing the date generated and the go release used, for example: For example, the homeobox d9a gene product from zebrafish has numerous go terms associated with it.
This site is an open community for users to do submittion their favorite wallpapers on the internet, all images or pictures in this website are for personal wallpaper use only, it is stricly prohibited to use this wallpaper for commercial purposes, if you are the author and find this image is shared without your permission, please kindly raise a DMCA report to Us.
If you find this site beneficial, please support us by sharing this posts to your preference social media accounts like Facebook, Instagram and so on or you can also bookmark this blog page with the title gene ontology citation by using Ctrl + D for devices a laptop with a Windows operating system or Command + D for laptops with an Apple operating system. If you use a smartphone, you can also use the drawer menu of the browser you are using. Whether it’s a Windows, Mac, iOS or Android operating system, you will still be able to bookmark this website.
Category
Related By Category
- De vliegeraar citaten information
- Full reference citation apa style information
- Free apa citation machine online information
- Etre amoureux citation information
- Fight club citation tyler information
- Evene lefigaro fr citations information
- Freud citations aimer et travailler information
- Endnote book citation information
- Flap lever cessna citation information
- Foreign aid debate citation information